Next Generation Sequencing Data Science

Covid-19 RNA Seq Analysis
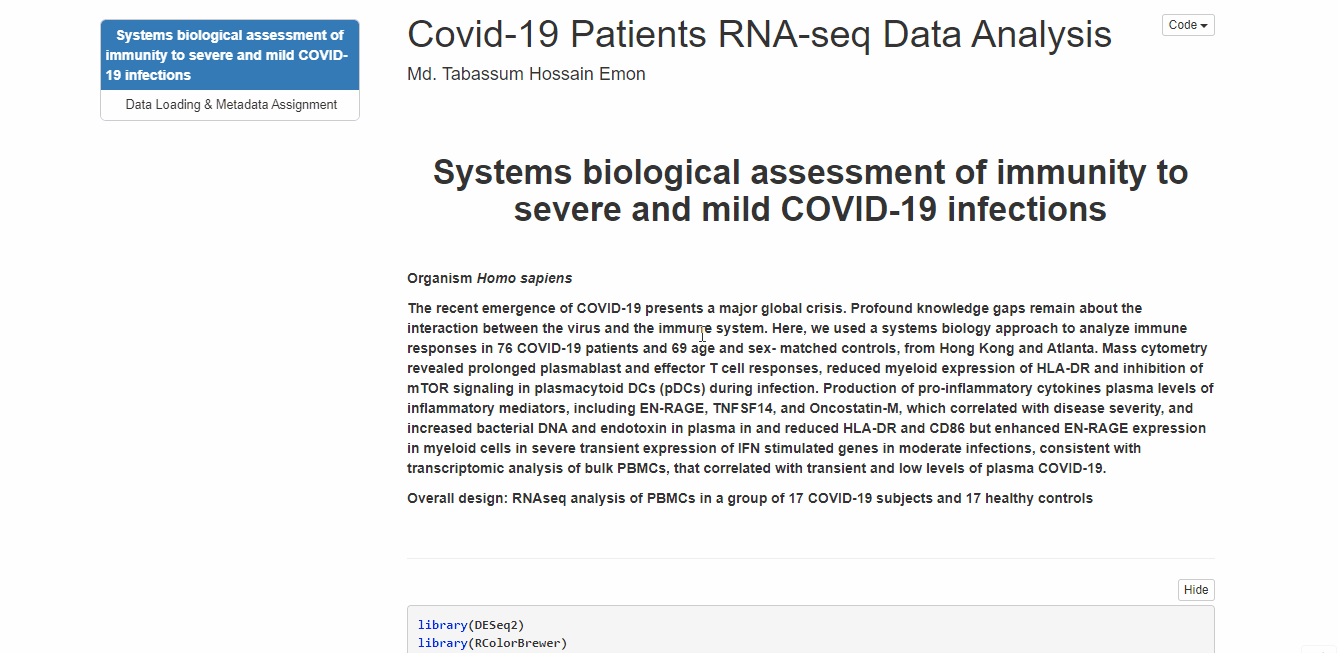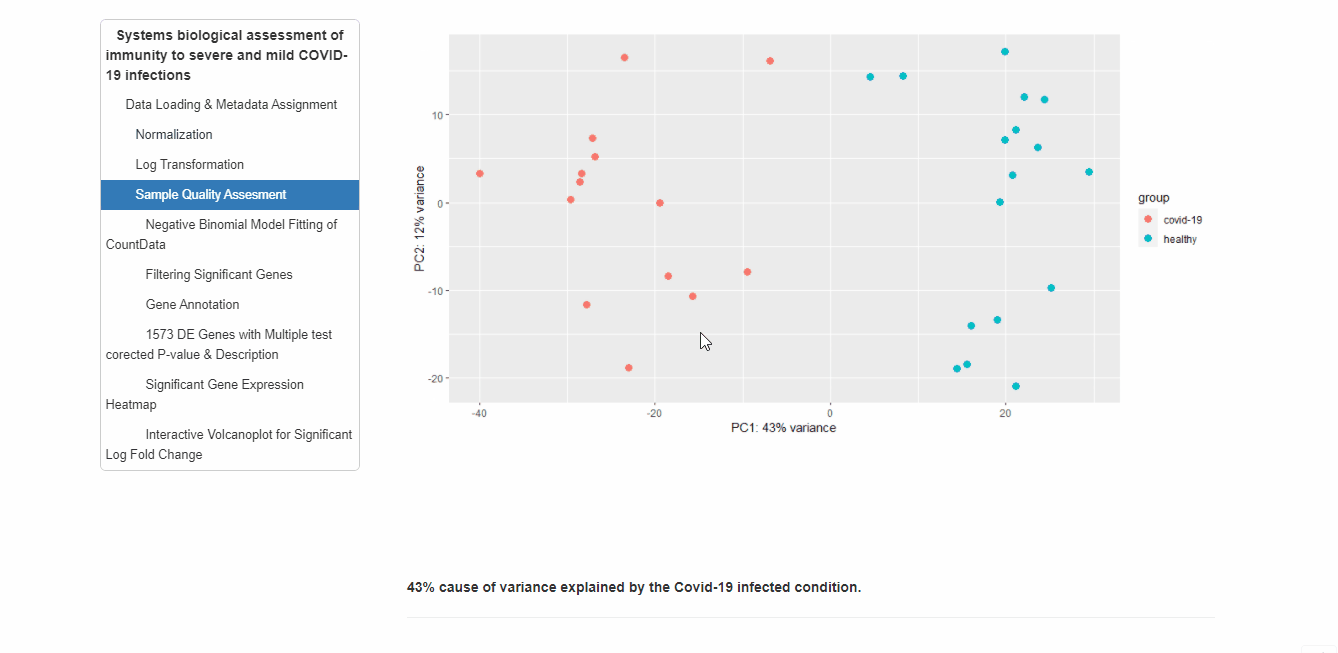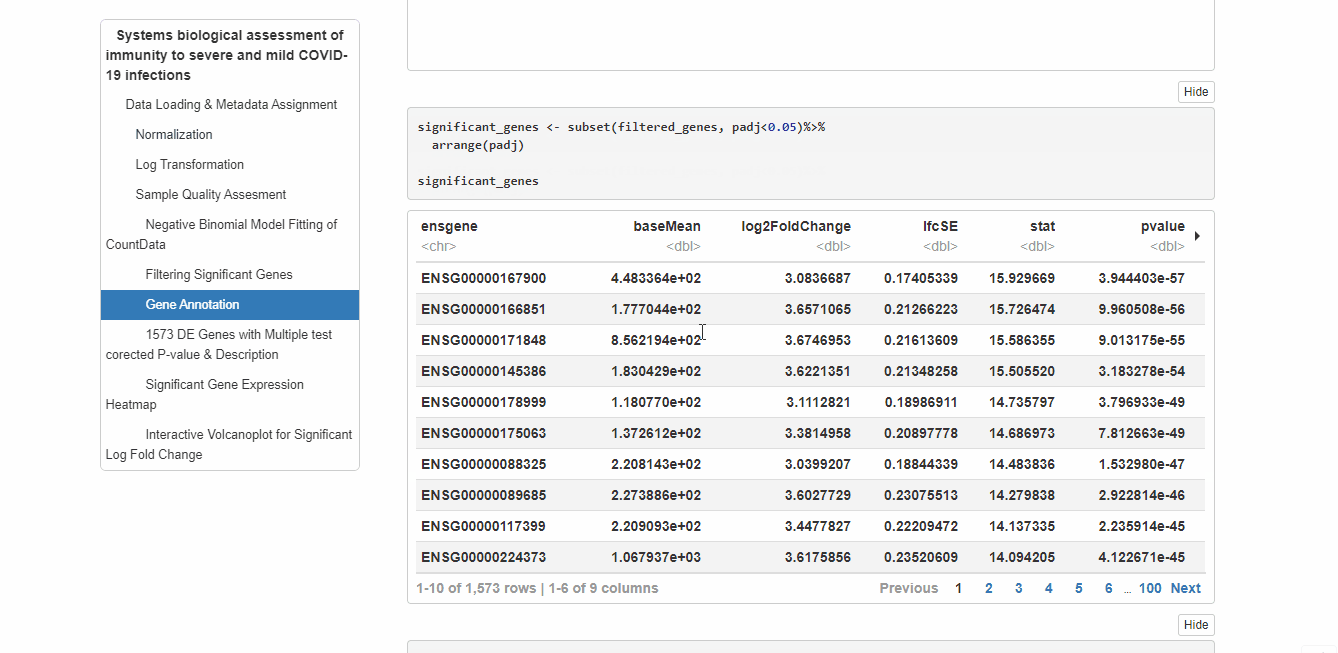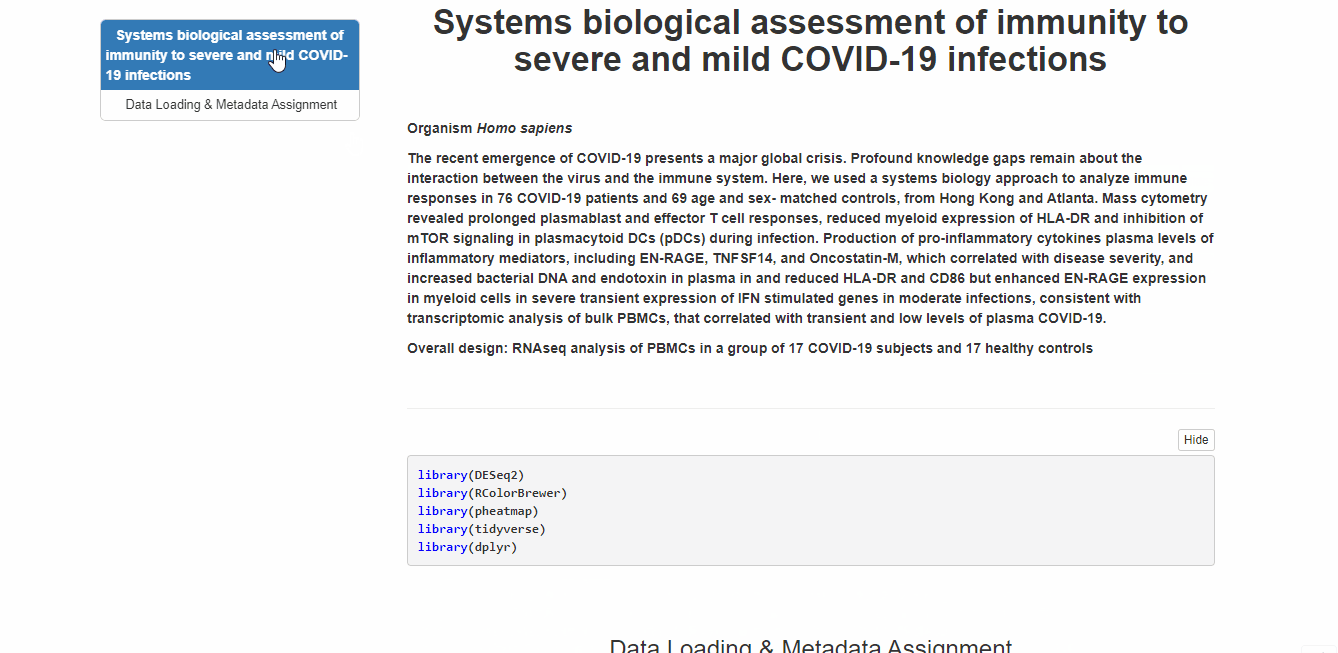
RNAseq analysis of PBMCs in a group of 17 COVID-19 subjects and 17 healthy controls. I have used DESeq2 to analyze this data. DESeq has been developed to enable analysis of experiments with small numbers of replicates. With DESeq, it is technically possible, although not recommended, to work with experiments without any biological replicates.
- Data Source: GSE-152418
👆🏻 Click The Image
Mechanism of Doxorubicin-induced Cardiotoxicity with limma
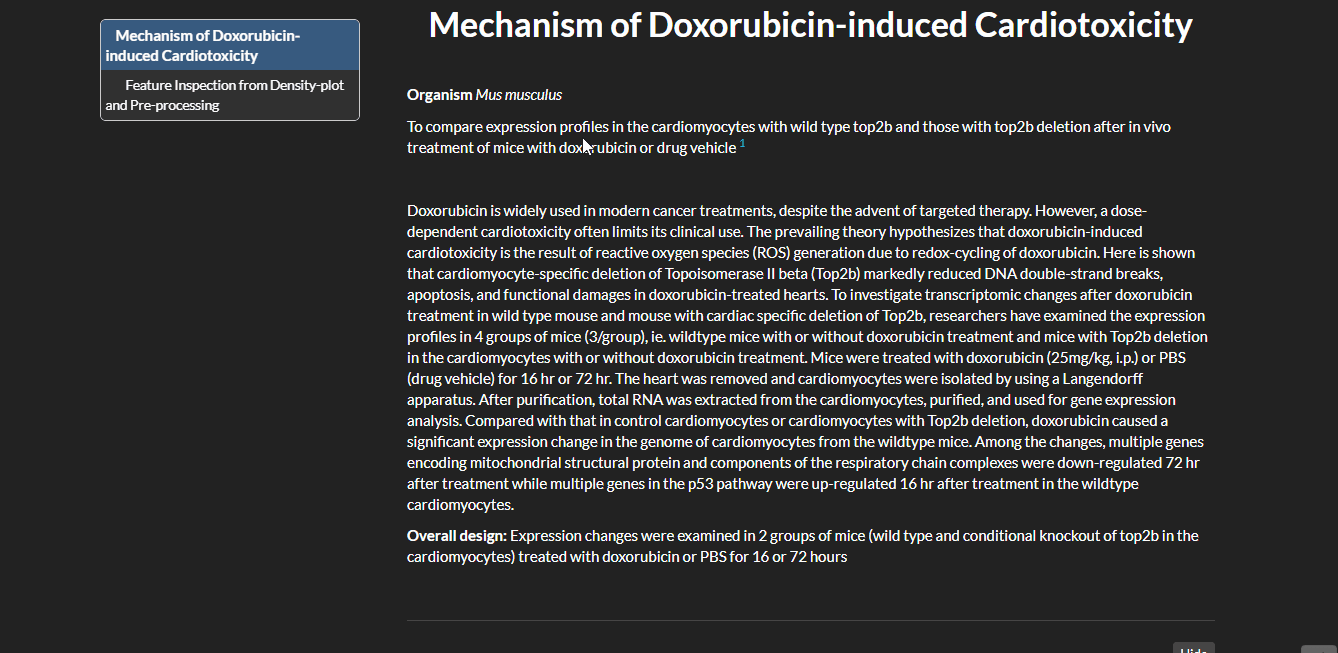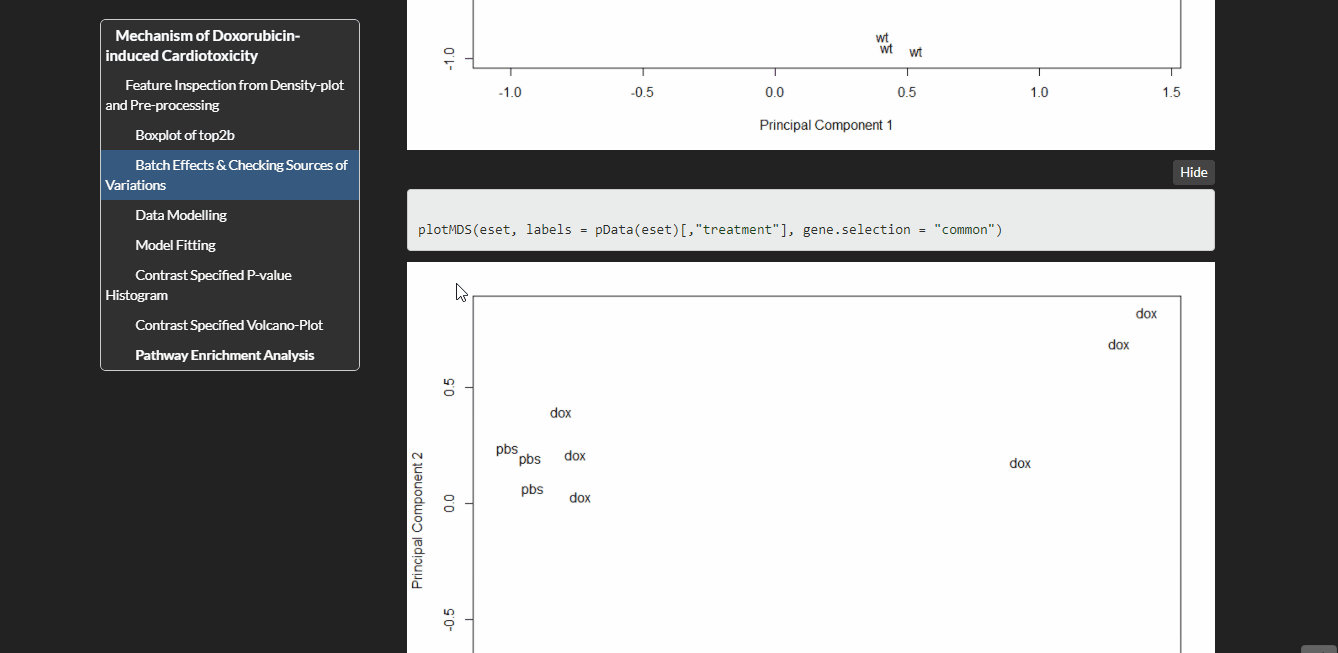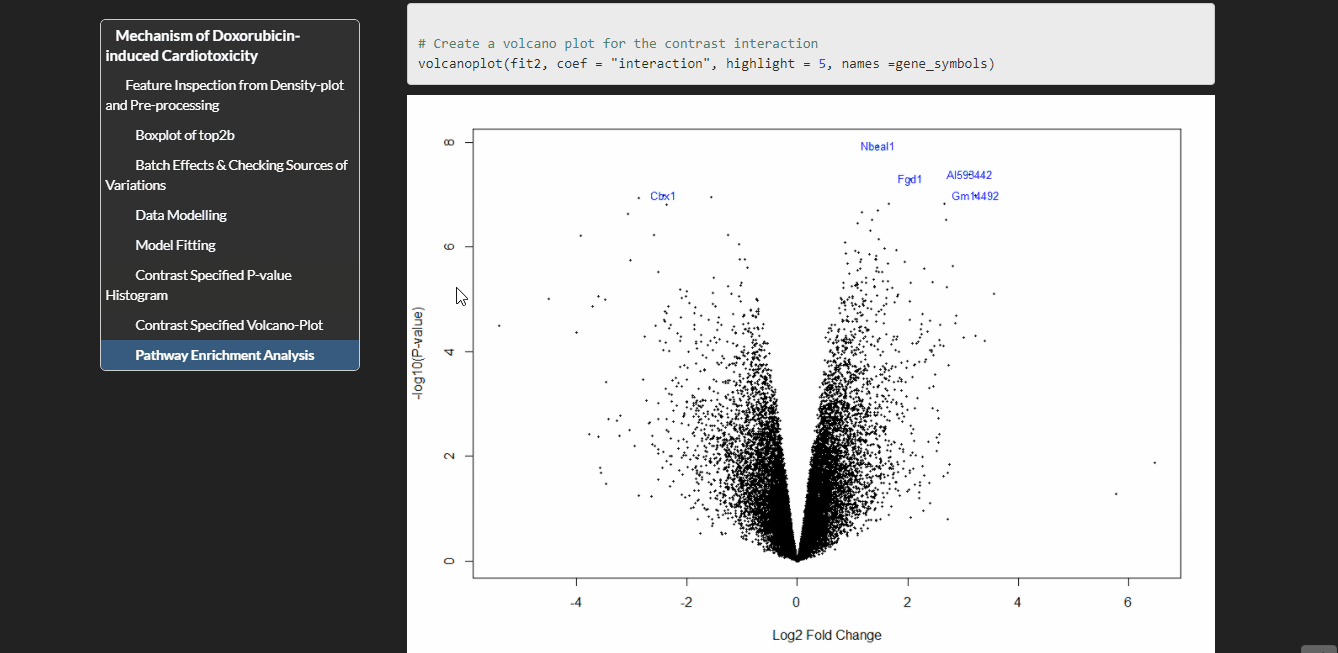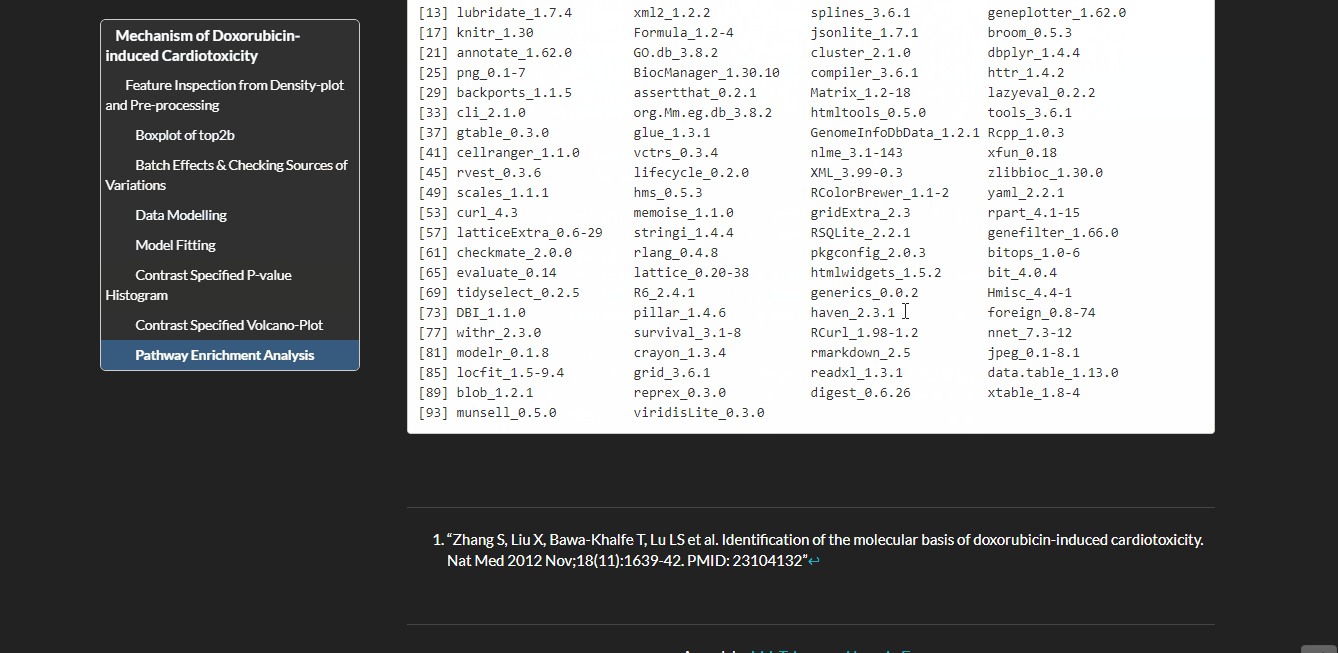
Expression changes were examined in 2 groups of mice (wild type and conditional knockout of top2b in the cardiomyocytes) treated with doxorubicin or PBS for 16 or 72 hours. I have used Limma package to analyze this data. Limma is based on linear modeling. It was originally designed for analyzing microarray data but has been extended to RNA-seq data.
- Data Source: GSE-40289
👆🏻 Click The Image
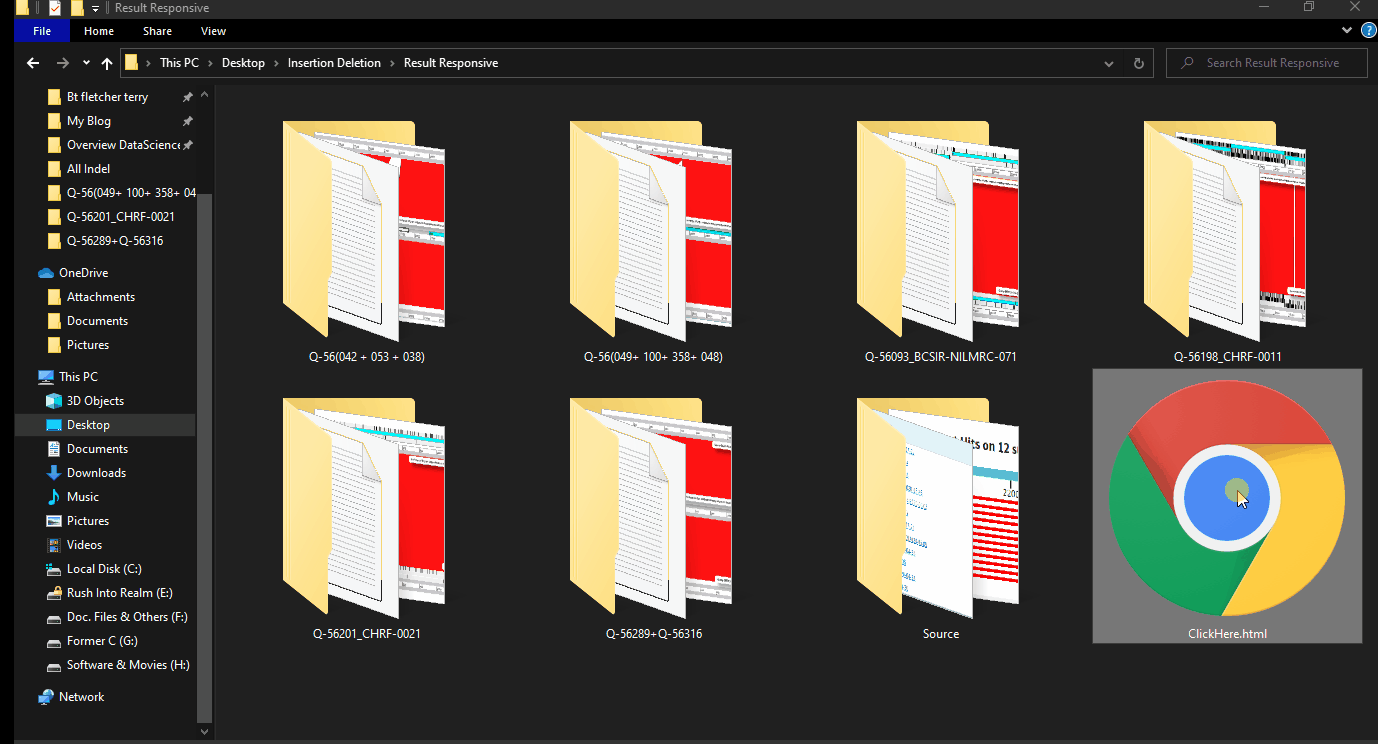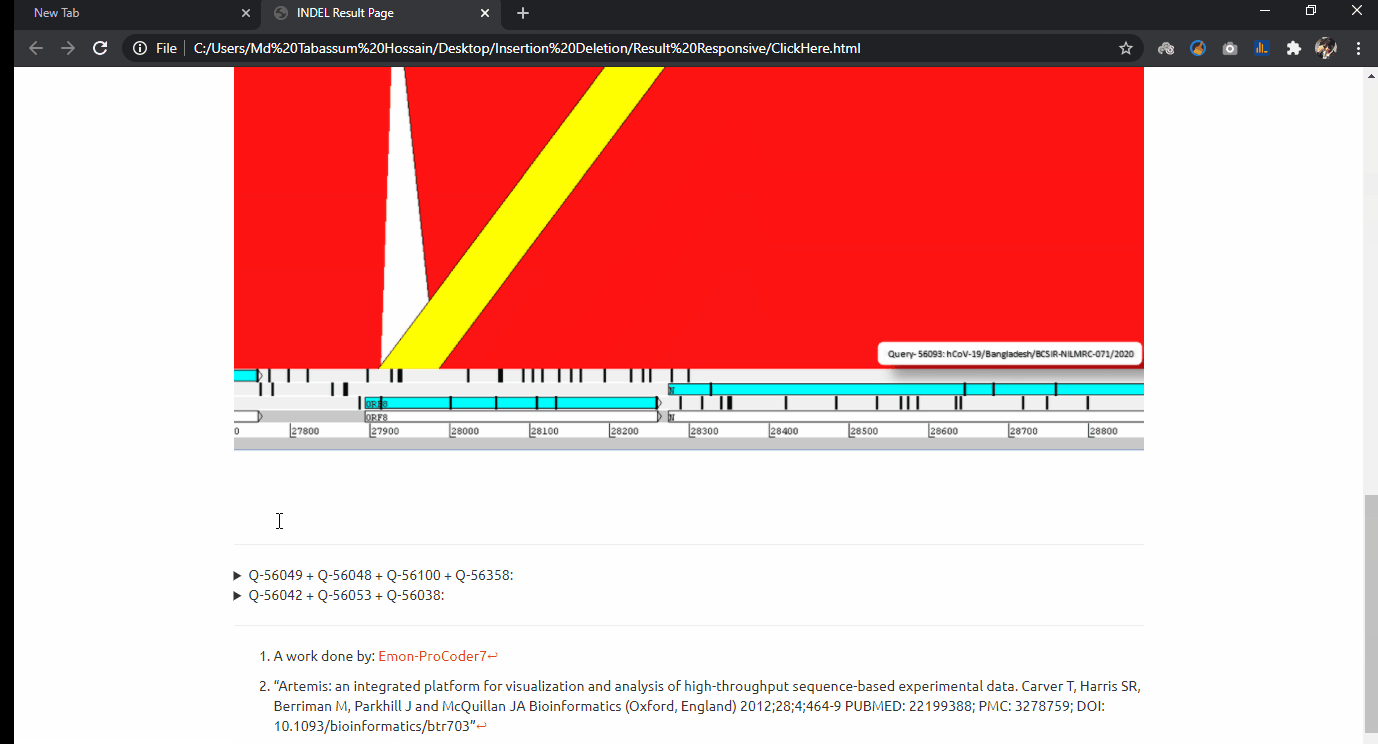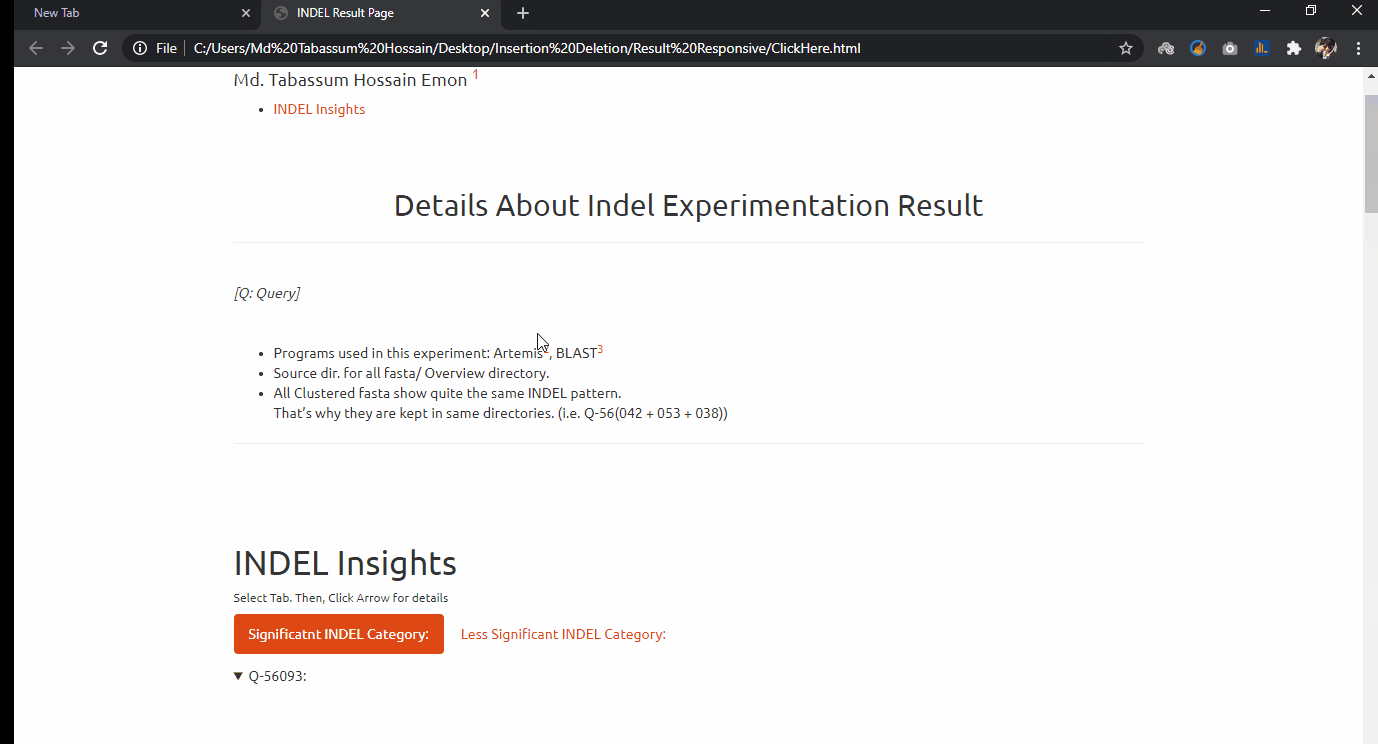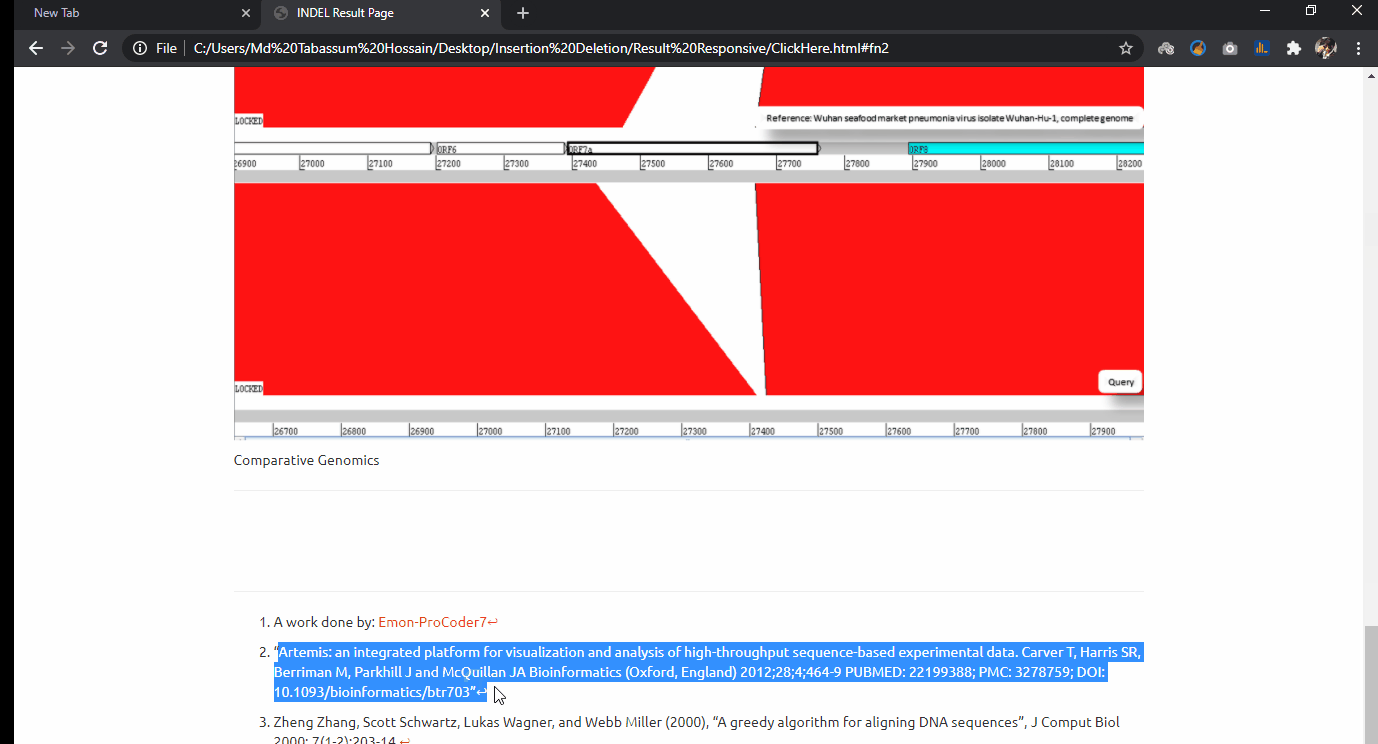
INDEL Analytics on Bulk Covid-19 Sequence Data
Indel experimentation result from comparative genome browsing against reference genome. Last updated on 11 October, 2020.
- Experimentation on: 323 Sequenced Sars CoV-2 genomes up to date in Bangladesh.
- Project Goal: From the massive sequence data alignment, I have sorted out these genomes and made comparative INDEL visualization in a genome browsing window.
On the result page, Select Tab. Then, Click ▶ button for details