Pathogenic Genetic Variants From Highly Connected Cancer Susceptibility Genes Confer the Loss of Structural Stability
Abstract
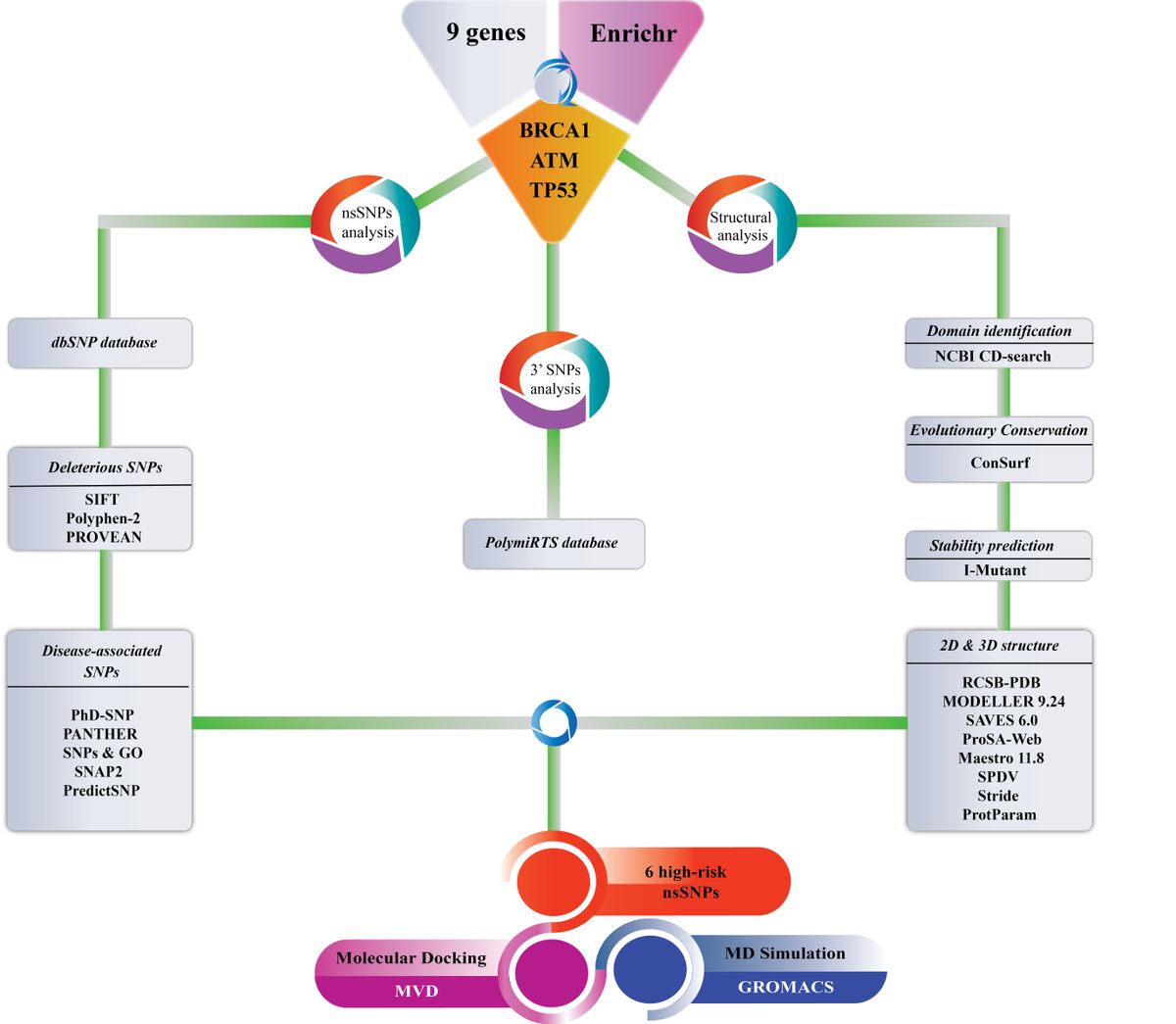
Genetic polymorphisms in DNA damage repair and tumor suppressor genes have been associated with increasing the risk of several types of cancer. Analyses of putative functional single nucleotide polymorphisms (SNP) in such genes can greatly improve human health by guiding choice of therapeutics. In this study, we selected nine genes responsible for various cancer types for gene enrichment analysis and found that BRCA1, ATM, and TP53 were more enriched in connectivity. Therefore, we used different computational algorithms to classify the nonsynonymous SNPs which are deleterious to the structure and/or function of these three proteins. Our study demonstrated that V1687G and V1736G variants of BRCA1, I2865T and V2906A variants of ATM, V216G and L194H variants of TP53 are major mutations with pathogenic impact and are likely to have a greater impact on destabilizing the proteins. To stabilize the high-risk SNPs, we performed mutation site-specific molecular docking analysis and validated using molecular dynamics (MD) simulation and molecular mechanics/Poisson Boltzmann surface area (MM/PBSA) studies. Additionally, SNPs of untranslated regions of these genes affecting miRNA binding were characterized. Hence, this study will assist in developing precision medicines for cancer types related to these polymorphisms.