 Thesis Outline
Thesis OutlineTAILORED BIOINFORMATICS & MACHINE LEARNING APPROACH FOR HACKING INTO COVID-19 MICROBIAL SYSTEM
Abstract
The human microbiome is a very active area of research due to its potential to explain health and disease. Advances in high throughput DNA sequencing have catalyzed the growth of microbiome research; DNA sequencing allows for an effective method to characterize entire microbial communities directly, including unculturable microbes which were previously difficult to study. Shotgun metagenomics and 16S rRNA sequencing, coupled with bioinformatics methods have powered the characterization of the human microbiome in different parts of the body. This has led to the discovery of novel links between the microbiome and diseases such as allergies, cancer, and autoimmune diseases.
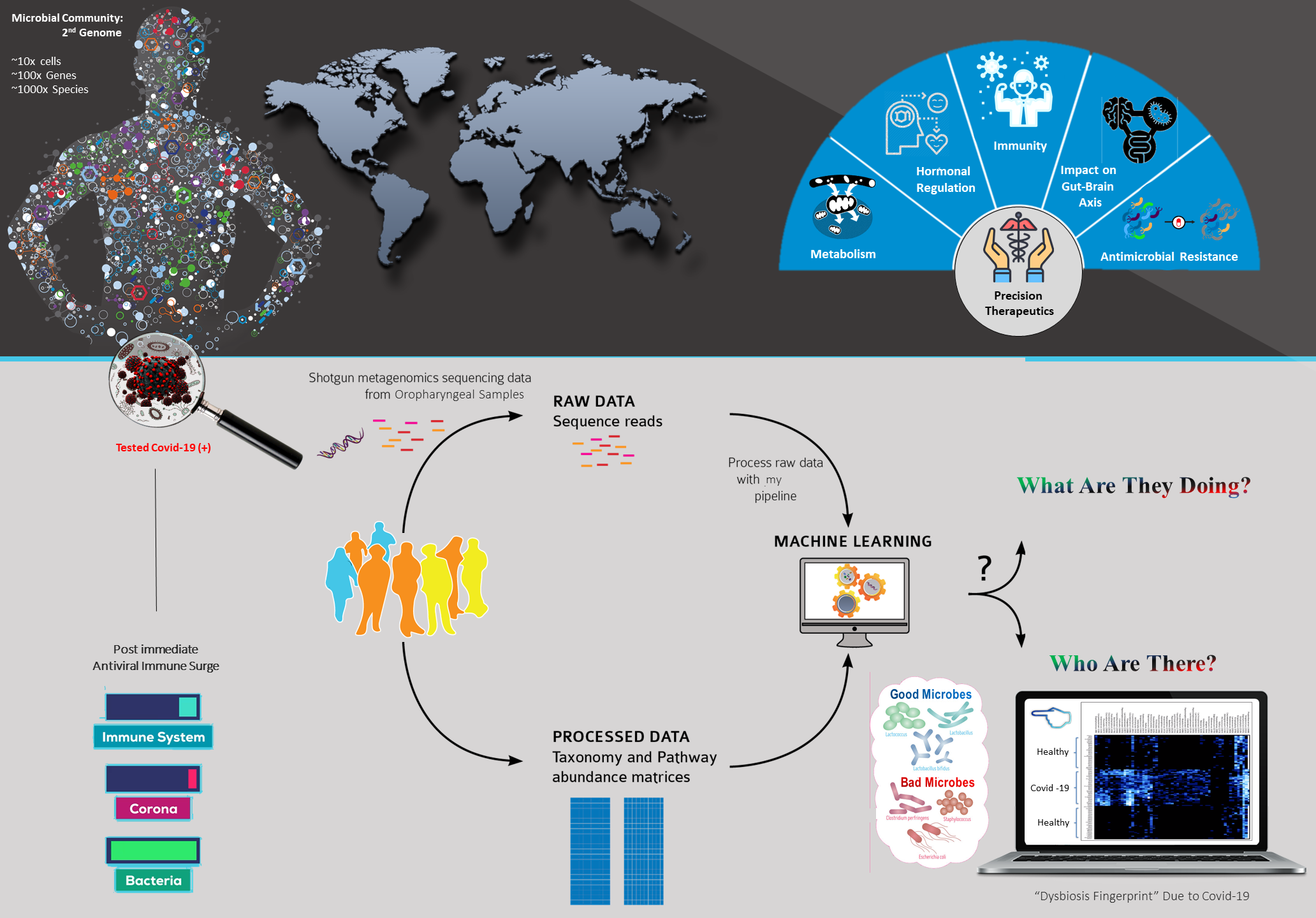
The rapid epidemic emergence of Covid-19 has devastated human-kind. For rapidly emerging infectious disease like Covid-19, metagenomic next-generation sequencing (mNGS) offers an opportunity to both recover whole viral genomes for epidemiological purposes and to agnostically determine coinfections that may be associated with increased morbidity and mortality. Particularly, Whole Genome mNGS approach could simultaneously reveal the entire “infectome” (i.e. RNA viruses, DNA viruses, bacteria and eukaryotes) present within an individual.
This thesis is part of the BCSIR-NIB mNGS collaboration project, which focuses on the application of shotgun metagenomics for the characterization of the Oropharyngeal sample collected from Covid-19 patient. I have used Machine Learning methodologies supported by cloud computing, for breaking into Covid-19 Microbial System to answer the two-classic question of metagenomics- “Who is there?” & “What are they doing?”. So that, this taxonomical and functional data can be merged with comprehensive patient profiles, including demographics and family history; traditional laboratory data; and next-generation ‘omics’ data for accelerated precision therapeutics with maximal efficacy.